Yes you’re right I realized it didn’t work
.count 66145.000000
mean 1.641974
std 0.067499
min 1.500000
25% 1.590000
50% 1.650000
75% 1.690000
max 1.790000
Name: height, dtype: float64
count 66465.000000
mean 73.730712
std 11.904887
min 51.500000
25% 65.000000
50% 72.000000
75% 81.000000
max 107.000000
Name: weight, dtype: float64```I don’t understand why it still doesn’t work if I’ve now fixed it as you said
Can you show the .describe() before and after?
Minimum height looks ok
Another problem is this line:
df_heat = df.copy()
df_heat['ap_lo']=df_heat['ap_lo'][df_heat['ap_lo']<=df_heat['ap_hi']]
print(df_heat['ap_lo'])
print(df['ap_lo'])
df_heat['ap_lo'] is unchanged after the line is run
0 80.0
1 90.0
2 70.0
3 100.0
4 60.0
...
69995 80.0
69996 90.0
69997 90.0
69998 80.0
69999 80.0
Name: ap_lo, Length: 70000, dtype: float64
0 80
1 90
2 70
3 100
4 60
...
69995 80
69996 90
69997 90
69998 80
69999 80
Name: ap_lo, Length: 70000, dtype: int64
It seems like you will need to filter into a new dataframe
https://www.geeksforgeeks.org/ways-to-filter-pandas-dataframe-by-column-values/
You could combine these two lines:
df_heat = df.copy()
df_heat['ap_lo']=df_heat['ap_lo'][df_heat['ap_lo']<=df_heat['ap_hi']]
Instead of making a copy into df_heat first, you can filter df into the new dataframe df_heat
ok i will try it and update you. thanks for your help really and sorry!
I get this error as I told you when I try to convert float to int and I used copies for it first
raise IntCastingNaNError(
pandas.errors.IntCastingNaNError: Cannot convert non-finite values (NA or inf) to integer
Anyway, now I’m looking for a solution to that error but now I’ve lost hope that it will be able to solve the heatmap error.
df_heat=df.copy()
df_heat['ap_lo']=df['ap_lo'][df['ap_lo']<=df['ap_hi']]
i will try later and update you
It’s not the float/int that’s a problem here but that it remains 70,000 lines and nothing is filtered out. I fixed it using an intermediate step for some reason filtering didn’t work “in place”
The note about the heights isn’t a float/int problem (although that might be something else to look at)
Height is supposed to be in centimetres. Numbers seem quite small, right?
Yes, you’re right. However, it’s true that filtering doesn’t work as it should and I noticed it. I have to see if I can use some intermediate steps too. Thanks for the really help and sorry for the inconvenience.
It’s no problem, I’m happy to help. We’re almost there!
Yes, later I’ll try to see how to filter more effectively. Yes, we’re close!
I checked the filters and they all work even without intermediate passage. so the problem has to be elsewhere. However in my opinion it could be that being then the columns in float after the filter and should be in int does not recognize them.
Then I managed to convert to int ‘ap_lo’ and ‘height’ after a few steps but I noticed that when I convert height back to cm I pass it again as float
ap_lo looks good:
count 70000.000000
min -70.000000
max 11000.000000
Name: ap_lo, dtype: float64
count 68766.000000
min -70.000000
max 182.000000
Name: ap_lo, dtype: float64
Not sure why that int conversion isn’t working but if you print it right after loading the csv it starts a float so I wouldn’t worry about that for now.
df_heat['height']=df_heat['height'].apply(lambda c:c*100)
df_heat['height']=df_heat['height'].astype(int)
df_heat['height']=df_heat['height'].apply(lambda h:h/100)
You convert height back to cm here, but then undo it by dividing by 100 again, not sure what you’re trying to accomplish here?
0 1.68
1 1.56
2 1.65
3 1.69
4 1.56
...
69994 1.65
69995 1.68
69996 1.58
69998 1.63
69999 1.70
Name: height, Length: 65000, dtype: float64
Height of 1.65 cm doesn’t make sense.
You also try to convert it back to cm after your quantile calculations, not before, but it doesn’t seem to affect it. I would just convert it back to cm immediately after calculating overweight, but it doesn’t make a difference for the quantile calculations.
The main problem is this does not give the correct result:
df_heat['height']=df_heat['height'][df_heat['height']>=quantile_1]
The quantile is correct. This format of filtering, something is wrong with it. It’s also inconsistent:
df_heat['height']=df_heat['height'][df_heat['height']>=quantile_1]
df_heat['height']=df_heat['height'][df_heat['height']<quantile_2]
df_heat['weight']=df_heat['weight'][df_heat['weight']>quantile_weight]
df_heat['weight']=df_heat['weight'][df_heat['weight']<quantile_weight_1]
Sometimes you do >= sometimes it’s >, why?
They give you the filter in the instructions:
height is less than the 2.5th percentile
Keep the correct data with
(df['height'] >= df['height'].quantile(0.025))
To use the filter is like this:
filtered = df[filter]
or
df = df[df['height'] >= df['height'].quantile(0.025)]
This filters the dataframe according to the boolean of the filter. This part of the video might help:
https://youtu.be/GPVsHOlRBBI?t=16625
To illustrate a bit futher here is an example dataframe:
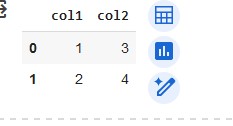
The way you were filtering:
df = df['col1'][df['col1']>1]

Result doesn’t quite make sense… if you wanted col1 where col1 is > 1
Correct way:
df = df[df['col1']>1]

The resulting dataframe only has rows where col1 was > 1
So, ap_lo is good, all you need to do is apply this filter format for each quantile here:
- height is less than the 2.5th percentile (Keep the correct data with
(df['height'] >= df['height'].quantile(0.025)))- height is more than the 97.5th percentile
- weight is less than the 2.5th percentile
- weight is more than the 97.5th percentile
It should be 4 lines of code, total.
Wait until I read everything. anyway I realized that even ‘gluc’ and ‘cholesterol’ have a problem maybe. because with map I change to 1 if it is greater than 1 otherwise 0 but there could also be the case where it is less than 1 and I’m replacing now
Then I converted them height to int immediately after filtering but before converting it to int I converted it into meters and then again into cm. Anyway I realized that it does not filter as it should with regard to the first quartile and now I’m trying your solution.
